Abstract
Clonal hematopoiesis (CH) in the elderly is associated with increased risk for a variety of diseases, including myeloid malignancies. However, as not all CH subjects develop myeloid malignancy, there is heterogeneity in clinical outcome. Indeed, longitudinal studies over several years have shown that clonal growth rates vary considerably between individual cases, even for the same CH driver mutation. In particular, with increasing age, clones may become large not only by positive selection, but also stochastically through neutral drift. Thus, personalized diagnostics are needed to evaluate evolution of CH.
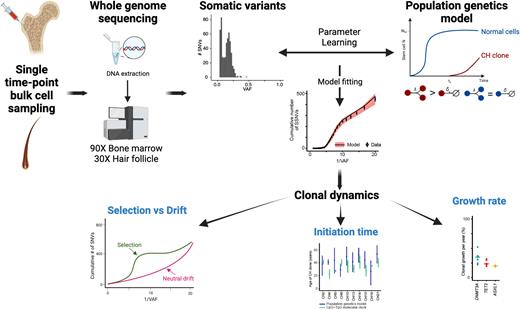
We analyzed the dynamics of normal and clonal hematopoiesis in twenty-one individuals with driver mutations in clonal hematopoiesis (CH)-associated genes and ten normal controls. To this end, we devised a population genetics model that learns the dynamics of normal and clonal hematopoiesis from the somatic variant allele frequency distribution in a single-patient bulk tissue sample. Moreover, the model can distinguish between drift and selection, both of which lead to somatic mosaicism but with different consequences for clonal evolution. To calibrate our model on human hematopoiesis, we identified somatic variants from deep (90X) whole-genome sequencing (WGS) of bulk bone marrow samples from our cohort and thereafter learned the model parameters using approximate Bayesian computation.
We found that clones with CH driver mutations originate decades before they are detected and confirmed these results using an independent approach that is based on CpG>TpG molecular clock timing. Thereafter, they expand due to positive selection rather than drift, with a selective advantage corresponding to a median growth rate of 26% per year in clones with a VAF of greater than 5%. While the selective advantage varied substantially between individuals, on average, DNMT3A variants conferred the strongest advantage, followed by TET2 and then ASXL1. By contrast, we did not detect a clear signal for clonal selection in bone marrow samples lacking known CH drivers. Remarkably, even CH clones with large advantage and growing over several decades did not make up the entire bone marrow in any of the analyzed individuals suggesting these clones grow with non-linear rates.
In summary, we provide a time-efficient and cost-effective method to study clonal dynamics of hematopoiesis at a large scale, which, by reducing cost and time, democratizes the research of somatic mosaicism and can be applied to other tissues. In addition, given that this method can reliably estimate clonal dynamics parameters such as time of initiation and growth rate, it may well serve current efforts in early detection of people at high risk for aggressive malignancy resulting from rapid clonal growth in a personalized medicine setting.
Disclosures
Vyas:Astellas: Honoraria; Celgene: Honoraria, Research Funding; Abbvie: Honoraria; Bristol Myers Squibb: Research Funding; Daiichi Sankyo: Honoraria; JAZZ: Honoraria; Pfizer: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.